Tracking Deadly Superbug Infections Across Europe with Web-Based Open Tools that Use Genome Sequencing and Open APIs
The Centre for Genomic Pathogen Surveillance has developed a new web-based tool that can track the spread of MRSA
 For the first time, scientists have shown that MRSA (methicillin-resistant Staphylococcus aureus) and other antibiotic-resistant ‘superbug’ infections can be tracked across Europe by combining whole-genome sequencing with a web-based system. In mBio today (5 May 2016) researchers at Imperial College London and the Wellcome Trust Sanger Institute worked with a European network representing doctors in 450 hospitals in 25 countries to successfully interpret and visualise the spread of drug-resistant MRSA.
For the first time, scientists have shown that MRSA (methicillin-resistant Staphylococcus aureus) and other antibiotic-resistant ‘superbug’ infections can be tracked across Europe by combining whole-genome sequencing with a web-based system. In mBio today (5 May 2016) researchers at Imperial College London and the Wellcome Trust Sanger Institute worked with a European network representing doctors in 450 hospitals in 25 countries to successfully interpret and visualise the spread of drug-resistant MRSA.
MRSA and other superbugs are a life-threatening problem for all hospitals across Europe with an estimated 400,000 cases per year and 25,000 deaths from resistant, hospital-acquired infections.
To enable infection control teams across Europe to easily share information and to form a dynamic picture of the rise and spread of antibiotic-resistant bacteria, the scientists from the newly formed Centre for Genomic Pathogen Surveillance developed Microreact.org, a web-based visualisation and mapping tool.
“Drug resistance is a growing problem both in Europe and across the world and doctors need fast and accurate information to stop epidemics. Our study demonstrates the potential for combining whole-genome sequencing with internet-based visualisation tools to enable public health workers and doctors to see how an epidemic is spreading and make swift decisions to end it.”
Dr David Aanensen, head of the Centre for Genomic Pathogen Surveillance and joint lead author on the paper
The research team read the whole genomes of S. aureus samples to identify which bugs are related to each other, and which are resistant to antibiotics. Using this approach, the scientists were not only able to show the rise and spread of MRSA across Europe, but also provide a quicker way to identify new hotspots of resistance.
 Professor Hajo Grundmann“One of the problems is that these bacteria not only spread within and between hospitals, but they also change their genetic properties due to evolutionary processes over time. Microreact.org allows us to look at their evolution within the context of how they are spreading across Europe.”
Professor Hajo Grundmann“One of the problems is that these bacteria not only spread within and between hospitals, but they also change their genetic properties due to evolutionary processes over time. Microreact.org allows us to look at their evolution within the context of how they are spreading across Europe.”
Professor Hajo Grundmann, principal investigator on the study and Head of the Institute of Infection Prevention and Hospital Hygiene at the University Medical Centre Freiburg in Germany
In the paper, the scientists show that combining the drug-resistance profile of a bacteria with its whole-genome DNA sequence allowed them to build up a series of drug-resistance ‘DNA photofits’ for resistance to specific drugs. In the future, such an approach may help doctors decide on the best treatments more quickly and help bring drug-resistant outbreaks to an end.
 Professor Ed Feil“We’ve developed user-friendly analysis software that demonstrates how whole genome sequence data can be a powerful tool for pan-European surveillance of MRSA and other important pathogens. Being able to track the spread of outbreaks across the whole continent allows policymakers to identify potential risks to public health and implement appropriate prevention and control strategies.”
Professor Ed Feil“We’ve developed user-friendly analysis software that demonstrates how whole genome sequence data can be a powerful tool for pan-European surveillance of MRSA and other important pathogens. Being able to track the spread of outbreaks across the whole continent allows policymakers to identify potential risks to public health and implement appropriate prevention and control strategies.”
Professor Ed Feil, joint lead author from the Milner Centre for Evolution, at the University of Bath
Notes
Publication
Aanensen D et al. (2016) Whole genome sequencing for routine pathogen surveillance in public health: A population snapshot of invasive Staphylococcus aureus in Europe. mBio Published on 5 May 2016.
Infection statistics taken from
World Health Organization (WHO) - http://www.euro.who.int/en/health-topics/disease-prevention/antimicrobial-resistance/data-and-statistics
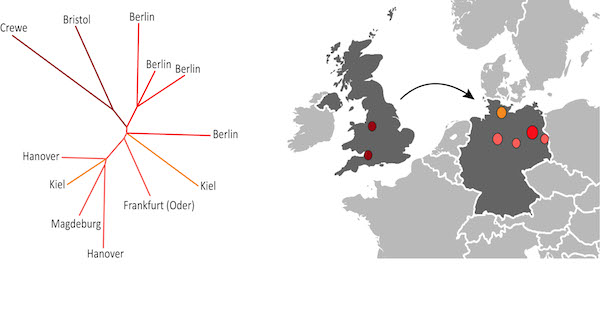 MRSA mapping in Europe - Credit David Anensen
MRSA mapping in Europe - Credit David Anensen
Participating Centres
- Department of Infectious Disease Epidemiology, School of Public Health, Imperial College London, London, United Kingdom
- The Centre for Genomic Pathogen Surveillance, Wellcome Genome Campus, Hinxton, Cambridge, United Kingdom
- The Milner Centre for Evolution, Department of Biology and Biochemistry, University of Bath, Bath, United Kingdom
- School of Medicine, University of St. Andrews, St. Andrews, United Kingdom
- Pathogen Genomics, The Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, United Kingdom
- Department of Biology, Drexel University, Philadelphia, Pennsylvania, USA
- Programa de Genómica Evolutiva, Centro de Ciencias Genómicas, Universidad Nacional Autónoma de Mexico, Cuernavaca, Morelos, Mexico
- Helsinki Institute for InformationTechnology HIIT, Aalto, Finland
- Department of Mathematics, Imperial College London, London, United Kingdom
- Department of Medical Microbiology, University Medical Center Groningen, Rijksuniversteit Groningen, Groningen, The Netherlands
- National Institute for Public Health and the Environment (RIVM), Bilthoven, The Netherlands
- Centre Hospitalier Universitaire de Nice, Nice, France EUCAST Development Laboratory, Växjö, Sweden
- Department of Infection Prevention and Hospital Hygiene, Faculty of Medicine, University of Freiburg, Freiburg, Germany
Funding
This work was funded by the Wellcome Trust, grants 098051, 099202 and 089472, and the Medical Research Council (MRC) grant G1000803.
Selected Websites
The Centre for Genomic Pathogen Surveillance
The Centre for Genomic Pathogen Surveillance is a joint initiative between Imperial College London and The Wellcome Trust Sanger Institute. The centre seeks to provide data and tools to allow researchers, doctors and governments worldwide to track and analyse the spread of pathogens and antimicrobial resistance.
The University Medical Centre, Freiburg
The University Medical Centre is among the largest university medical centres in Germany and engages in research, teaching, and healthcare as its core responsibilities. Experts in a broad range of fields work together in an interdisciplinary environment to offer our patients optimal treatment informed by the latest scientific findings. The Faculty of Medicine of the University of Freiburg is one of the most renowned and highly decorated faculties in Germany. Thanks to the broad range of disciplines covered, 3.575 medical students receive virtually limitless opportunities to extend their knowledge close to patients. The University of Freiburg has been ranked among Germany’s top higher education institutions on the CHE University Ranking for many years.
Imperial College London is one of the world's leading universities. The College's 16,000 students and 8,000 staff are expanding the frontiers of knowledge in science, medicine, engineering and business, and translating their discoveries into benefits for society.Founded in 1907, Imperial builds on a distinguished past - having pioneered penicillin, holography and fibre optics - to shape the future. Imperial researchers work across disciplines to improve health and wellbeing, understand the natural world, engineer novel solutions and lead the data revolution. This blend of academic excellence and its real-world application feeds into Imperial's exceptional learning environment, where students participate in research to push the limits of their degrees.Imperial collaborates widely to achieve greater impact. It works with the NHS to improve healthcare in west London, is a leading partner in research and education within the European Union, and is the UK's number one research collaborator with China.Imperial has nine London campuses, including its White City Campus: a research and innovation centre that is in its initial stages of development in west London. At White City, researchers, businesses and higher education partners will co-locate to create value from ideas on a global scale.
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute is one of the world's leading genome centres. Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease.
The Wellcome Trust is a global charitable foundation dedicated to improving health. We support bright minds in science, the humanities and the social sciences, as well as education, public engagement and the application of research to medicine. Our investment portfolio gives us the independence to support such transformative work as the sequencing and understanding of the human genome, research that established front-line drugs for malaria, and Wellcome Collection, our free venue for the incurably curious that explores medicine, life and art.
Contact the Press Office
Dr Samantha Wynne
Media Officer
Wellcome Trust Sanger Institute,
Hinxton,
Cambridgeshire,
CB10 1SA,
UK
Tel +44 (0)1223 492 368
Mobile +44 (0)7753 775 397
Fax +44 (0)1223 494 919
Email: [email protected]
- Tags:
- antibiotic-resistant infections
- Antibiotics
- antimicrobial resistance
- Centre for Genomic Pathogen Surveillance
- Centre Hospitalier Universitaire de Nice
- Centro de Ciencias Genómicas
- data sharing
- David Aanensen
- Department of Infectious Disease Epidemiology
- Drexel University
- drug resistance
- drug-resistance DNA photofits
- Ed Feil
- epidemic tracking
- EUCAST Development Laboratory
- Europe
- Hajo Grundmann
- Helsinki Institute for InformationTechnology HIIT
- hospital-acquired infections
- Imperial College London
- Imperial College London School of Public Health
- infection control
- Institute of Infection Prevention
- Medical Research Council (MRC)
- methicillin-resistant Staphylococcus aureus (MRSA)
- Microreact.org
- MRSA mapping in Europe
- National Institute for Public Health and the Environment (RIVM)
- Open Access
- open APIs
- open health
- Open Medicine
- Open Source APIs
- open source software (OSS)
- Pathogen Genomics
- Programa de Genómica Evolutiva
- public health workers
- superbug infections
- The Milner Centre for Evolution
- The Wellcome Trust Sanger Institute
- United Kingdom
- Universidad Nacional Autónoma de Mexico
- University Medical Center Groningen
- University Medical Centre Freiburg
- University of Bath
- University of Freiburg
- University of St. Andrews School of Medicine
- user-friendly analysis software
- web-based visualisation and mapping tool
- Wellcome Genome Campus
- Wellcome Trust Genome Campus
- Wellcome Trust Sanger Institute
- whole genome sequence data
- Login to post comments
