Peering into Complex, Tiny Structures with 3D Analysis Tool Tomviz
The future of technology is being built at the tiniest scales and now demands tools for 3D design and analysis.
 Robert HovdenNew open source software tomviz—short for tomographic visualization—enables researchers to interactively understand large 3D datasets. More specifically, the software analyzes 3D tomographic data similar to a medical CT-scan but at the nanoscale.
Robert HovdenNew open source software tomviz—short for tomographic visualization—enables researchers to interactively understand large 3D datasets. More specifically, the software analyzes 3D tomographic data similar to a medical CT-scan but at the nanoscale.
"When you can take a nanoparticle or biomolecule and spin it around, slice it, look inside it, and quantitatively analyze it, you get a complete picture from all angles," says Yi Jiang, a physics Ph.D. candidate at Cornell University.
Watch this 3-minute video from the Michigan Engineering department. Read the press release. For nanoscience, seeing in all dimensions is a big deal. The future of technology is being built at the tiniest scales and now demands tools for 3D design and analysis so that they can peer into complex structures. One only needs to look at Manhattan skyscrapers to appreciate the exponential benefit of 3D architecture—the largest city in America squeezed into a small island. Greater population densities are only possible due to the increased use of the third dimension to erect ever taller buildings that accommodate vast numbers on relatively small areas of land.
Your next computer processor will harness powerful 3D design to squeeze billions of nano devices onto a small chip. This is already beginning to happen in the memory market, where multiple layers of memory are stacked, and interconnects offer more memory bandwidth/storage density than possible before. Materials tomography is an enabling technique in the design, testing, and validation of 3D device structures that support further miniaturization.
tomviz 1.0 is new software for processing and visualizing 3D data, making it immediately popular among nanoscientists. "The algorithms we've developed in tomviz can handle big datasets and enable scientists to understand the nanoscale universe across dimensions," says Yi Jiang.
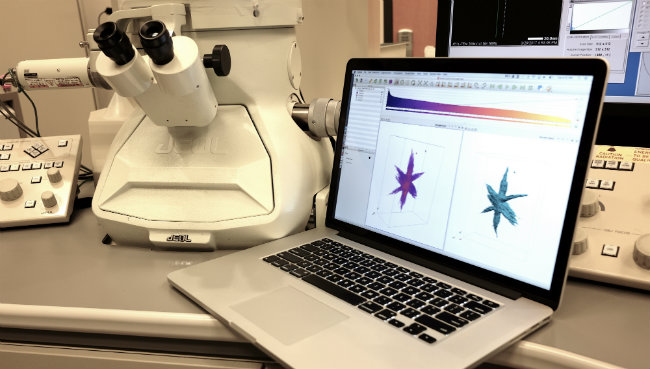 tomviz works with electron microscopes to provide 3D images of nanomaterials. (Robert Hovden - CC BY)Now scientists can rapidly study the nanoscale in 3D at the highest resolution by utilizing data acquired from an electron microscope and loading that data into the tomviz software. "With modern electron microscopes you can see the image of a single atom—it's incredible. But it's only a 2D image and jumping to 3D is an extremely complicated process. We've democratized this complex process with our software," says Dr. Peter Ercius, a Berkeley lab scientist. The entire data processing pipeline from raw data to visualization to statistical analysis is streamlined within tomviz 1.0.
tomviz works with electron microscopes to provide 3D images of nanomaterials. (Robert Hovden - CC BY)Now scientists can rapidly study the nanoscale in 3D at the highest resolution by utilizing data acquired from an electron microscope and loading that data into the tomviz software. "With modern electron microscopes you can see the image of a single atom—it's incredible. But it's only a 2D image and jumping to 3D is an extremely complicated process. We've democratized this complex process with our software," says Dr. Peter Ercius, a Berkeley lab scientist. The entire data processing pipeline from raw data to visualization to statistical analysis is streamlined within tomviz 1.0.
Better tools; all in one place.
Most strikingly, the visualizations in tomviz 1.0 are gorgeous. Intricate nanoparticles used in clean energy vehicles are visualized by researcher Elliot Padgett to determine particles inside or outside of a winding tube of graphite. The design and location of these nanoparticles directly affect the performance of future clean vehicles; this requires 3D analysis. Clean energy is just one example of many industries that are designing future technology using 3D nanomaterials.
3D science critically needs open tools to meet the requirements of reproducible science—the foundation of scientific philosophy. The tomviz team built a platform that firmly supports open science and pushes the boundaries of 3D data science. The platform facilitates researchers to develop their own Python algorithms to run on the 3D data with one click execution.
The recent 3D revolution spans many fields of research from biology, medicine, and materials science but all are bottlenecked by a lack of capable, open, and collaborative software tools. That problem now has a solution. "Today’s v1.0 release of tomviz is a significant milestone for the project," says scientific programmer, Dr. Marcus Hanwell at Kitware, "it provides a comprehensive product that propels scientists forward when working in three dimensions at the nanoscale."
The project has been developed openly from the very beginning, predominantly focused on a C++ code base, using the permissive 3-clause BSD license, on GitHub. It leverages the very best of open source, including Python, NumPy, SciPy, Qt, ITK, VTK, and ParaView. Some of these are developed for general-purpose applications, while others have a strong focus on scientific data, from processing it to visualizing it.
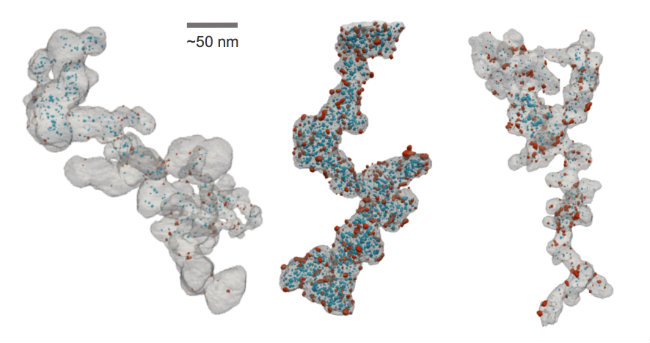 3D nanomaterials for clean-energy vehicles visualized in tomviz 1.0 by Elliot Padget (Cornell University). The key to performance depends on the particles inside (blue) or outside (red) the carbon structure (grey). (Elliot Padget - CC BY)The project has been designed with the research community in mind; adding new file formats to these toolkits to support getting data in and out of the application. Using public issue trackers, code review, and continuous integration to make the contribution process as transparent and collaborative as possible.
3D nanomaterials for clean-energy vehicles visualized in tomviz 1.0 by Elliot Padget (Cornell University). The key to performance depends on the particles inside (blue) or outside (red) the carbon structure (grey). (Elliot Padget - CC BY)The project has been designed with the research community in mind; adding new file formats to these toolkits to support getting data in and out of the application. Using public issue trackers, code review, and continuous integration to make the contribution process as transparent and collaborative as possible.
The data operations are primarily developed in Python and can be edited/executed right in the application. These are saved in the application state file, using an XML format, that can be restored and shared. The development of a data pipeline designed to be shared, saved, edited, and redistributed enables not only sharing open data but all the steps it takes to get there in an editable form never seen before in this area.
A nascent idea that began in the Hovden Lab, matured through a collaborative team spanning several institutions including the University of Michigan, Cornell University, Berkeley Lab, and Kitware. And because the tool is open source, a much greater community of talented developers has contributed.
The tomviz project serves not just as a tool for science, but as a model for open science software. Best of all, anyone can acquire, tomviz, today and explore the nanoworld.
| Peering into Complex, Tiny Structures with 3D Analysis Tool Tomviz was authored by Robert Hovden and published in Opensource.com. It is being republished by Open Health News under the terms of the Creative Commons Attribution-ShareAlike 4.0 International License (CC BY-SA 4.0). The original copy of the article can be found here. |
- Tags:
- 3-clause BSD license
- 3D architecture
- 3D data science
- 3D datasets
- 3D design and analysis
- 3D nanomaterials
- 3D tomographic data
- Berkeley Lab
- C++ code base
- code review
- collaboration
- Continuous Integration
- Cornell University
- Github
- Hovden Lab
- ITK
- Kitware
- Marcus Hanwell
- Michigan Engineering
- miniaturization
- nanoscience
- Numpy
- open source
- open source software (OSS)
- open source software tomviz
- ParaView
- Peter Ercius
- public issue trackers
- Python
- Qt
- Robert Hovden
- SciPy
- tomographic visualization (tomviz)
- tomviz 1.0
- transparency
- VTK
- Yi Jiang
- Login to post comments