Major German research project chooses openEHR
 Thomas Beale I just returned from Heidelberg, where another very successful ‘openEHR day’ was held, this time by the HiGHmed research consortium, with 100 attendees. HiGHmed is funded with 20m€ by the German Federal Ministry of Education and Research (BMBF) under the “Medical Informatics” funding scheme, and has as its goal (my bolding):
Thomas Beale I just returned from Heidelberg, where another very successful ‘openEHR day’ was held, this time by the HiGHmed research consortium, with 100 attendees. HiGHmed is funded with 20m€ by the German Federal Ministry of Education and Research (BMBF) under the “Medical Informatics” funding scheme, and has as its goal (my bolding):
.. to develop and use innovative information infrastructures to increase the efficiency of clinical research and to swiftly translate research results into validated improvements of patient care. These aims are tightly connected with challenges to integrate and further develop solutions of innovative, internationally interoperable data integration and methods, with the aim to demonstrate their added value for health research and patient care. The concepts must be designed in a way that will help to develop sustainable structures and with the perspective for an easy roll-out to other hospitals.
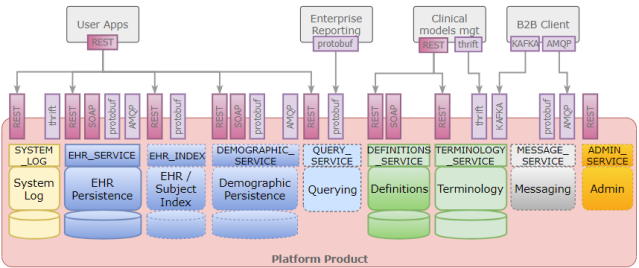
 The slide in the image above is from Wouter Zanen, who presented on EuroTransplant, an 8-country network that facilitates patient-oriented allocation and cross-border exchange of deceased donor organs, now using openEHR. But the reasons for HiGHmed’s choice are pretty close I think. A number of other presentations on openEHR deployments and projects say very similar things.
The slide in the image above is from Wouter Zanen, who presented on EuroTransplant, an 8-country network that facilitates patient-oriented allocation and cross-border exchange of deceased donor organs, now using openEHR. But the reasons for HiGHmed’s choice are pretty close I think. A number of other presentations on openEHR deployments and projects say very similar things.
One of the recurrent debates that surfaced during HighMed’s deliberations was the rerun of the now 18-year old question:
- why openEHR rather than FHIR (was CDA, was HL7v3)?
As you can see we are on the 3rd iteration of that particular conundrum. There are many long answers, provided by myself and others in the past, but the short version is roughly the same as I’ve been presenting for about 20 years now:
And the reality is that you can’t solve this with messages or documents or, as they are called now, ‘resources’. You can solve some very useful problems with all of these, particularly when the systems from which you are trying to get data are opaque. But the real question is: what about the system(s) you are putting the data into? That’s where you need a comprehensive platform architecture that is model-driven, semantically scalable and technology-neutral. You have to have things like:
- a proper formalism to do domain-level modelling (i.e. archetypes and templates) – we use BMM and ADL;
- a way to write model-based queries rather than schema-based queries – AQL, in openEHR;
- a way to generate message definitions for technology-independent interoperability.
- And you need a service architecture, and service APIs.
I’ve discussed some of this in previous posts – e-Health standards – beyond the message mentality; and FHIR compared to openEHR. Invariably when one asks: why do you think we should use FHIR? The response is usually something like: well that’s what everyone else is using. This was the case when the question was ‘why aren’t you using HL7v3?’. HL7v3 is now dead. FHIR is certainly far better than HL7v3, is implementable and well thought out in many respects. But the question that matters is: what do I need to solve my problem?
To provide a summary of what openEHR currently consists of:
- 28 specifications, with all UML models available online;
- 695 international archetypes (about 8,500 clinical data points);
- 42 GitHub repositories for software and other projects;
- O(100) deployments.
It is an ongoing process, not something complete, and generally progresses in the form of open source scientific research and development, rather than design-by-committee. At a purely technology level, openEHR is just a piece of an overall solution environment; you also need terminologies, possibly IHE services, you may need FHIR and/or other on-the-wire solutions, and you’ll need a whole slew of application and middleware technologies. But openEHR is also a methodological approach, possibly best summed up in the two overview documents openEHR Architecture overview, and the Archetype Technology overview. The shortest way to capture it I can think of is: semantics + platform as a process.
In my view, it would be much better to be able to cooperate more effectively with HL7 and make both types of technology work together rather than having dull arguments about how my orange is better than your apple. To make this possible, the FHIR-hype needs to disappear and be replaced by level-headed thinking (those involved for 10 years or more in e-health will know that 15 years of HL7v3 over-selling was not helpful to anyone, and cost a great deal of public and private money). That entails getting rid of the standards-as-a-competition mentality, being far more careful in raising expectations with major funding agencies and governments, and treating the journey to solutions as a scientific one, not a political one.
In my mind, we are here to solve the challenges of the hardest domain there is, for the benefit of society. The technologies we work on have only one aim: to create concrete improvements in the lives of real people, through improved healthcare and research.
One of the areas I think where openEHR could help HL7 / FHIR is to do with modelling domain content – what we call archetypes and templates, what FHIR calls profiles and extensions. The motivation is to get towards being properly domain model-driven, not message-driven. This was the goal of the CIMI effort, now an HL7 Technical Committee.
 In openEHR, we use Basic Meta-Model (BMM) and/or UML as the information layer, and ADL/AOM as the constraint model layer. The FHIR approach to representing profiles is to use something called StructureDefinition. Now, BMM+ADL/AOM has been around for 15 years, with tooling and is used ubiquitously these days (including by CIMI, since 2011), and I think implements substantially more semantics than StructureDefinition (although the latter may have some features that are not in BMM+ADL) – multi-level specialisation, composition, inheritance flattening and diff-ing and much else. If we find newer requirements, we can add them pretty quickly to the existing specifications and re-issue them.
In openEHR, we use Basic Meta-Model (BMM) and/or UML as the information layer, and ADL/AOM as the constraint model layer. The FHIR approach to representing profiles is to use something called StructureDefinition. Now, BMM+ADL/AOM has been around for 15 years, with tooling and is used ubiquitously these days (including by CIMI, since 2011), and I think implements substantially more semantics than StructureDefinition (although the latter may have some features that are not in BMM+ADL) – multi-level specialisation, composition, inheritance flattening and diff-ing and much else. If we find newer requirements, we can add them pretty quickly to the existing specifications and re-issue them.
Other areas that can be worked on together are terminology, which most likely requires openEHR adopting much of what FHIR has done, since it has faced the same reality of a world that isn’t just SNOMED CT but contains many terminologies, and solved many of the problems.
Progress on just these two items would potentially enable openEHR archetypes and templates to be used to machine-generate FHIR profiles. That would certainly be useful, including to HiGHmed and also various NHS projects in the UK.
Then we could get on to the really deep questions.
| Major German research project chooses openEHR was authored by Thomas Beale and published in Woland's cat. It is republished by Open Health News with permission from the author. The original copy of the article can be found here. |
- Tags:
- Archetype Technology
- archetypes
- Basic Meta-Model (BMM)
- CDA
- CIMI effort
- clinical research
- comprehensive platform architecture
- e-Health standards
- EuroTransplant
- FHIR
- German Federal Ministry of Education and Research (BMBF)
- German research project
- GitHub repositories
- health research
- HiGHmed
- HiGHmed research consortium
- HL7v3
- IHE services
- information infrastructures
- internationally interoperable data integration
- medical informatics
- modelling domain content
- National Health System (NHS)
- open health
- open source
- open source scientific research
- openEHR
- openEHR day
- patient care
- semantics
- service APIs
- service architecture
- sustainable structures
- technology-independent interoperability
- terminologies
- Thomas Beale
- UML models
- Woland's Cat
- Wouter Zanen
- Login to post comments